SimpleProx
OUr Platform Technology

Patented Technology: PCT/SG2020/050305
SimpleProx is a DNA barcoding technology that unlocks the power of DNA amplification via proximity activation which is target agnostic.
- DNA barcoding for non nucleic acid targets
- Specificity of affinity binder and sensitivity of DNA amplification
- Wash-free homogeneous assay format
- Rapid isothermal reaction
Multiple target types | Single Chemistry
Flexibly tailor the technology with affinity binders for a wide range of targets.

Key Benefits
PCR-like Sensitivity
Bringing femtomolar sensitivity for rapid protein test. No more trade-off between speed and accuracy.
Rapid
< 20-min sample-to-answer workflow which is > 75% faster than most immunoassays. Take actions now.
Streamlined Workflow
Our one-step workflow enables on-site testing at the point-of-need. Eliminate logistics worry.
Isothermal Reaction
Single temperature for the entire reaction. Simple to design and use.
High Reproducibility
Assay CV < 10% for precise test outcomes that you can trust.
Precise Quantification
Quantitative results which correlate with the biomarker level. Interpret disease severity.
Point-of-Care Testing (POCT) Application
SimpleProx offers lab quality test results with the speed and simplicity expected at the point-of-care. With our patented solution, there is no need to trade-off test accuracy for speed.
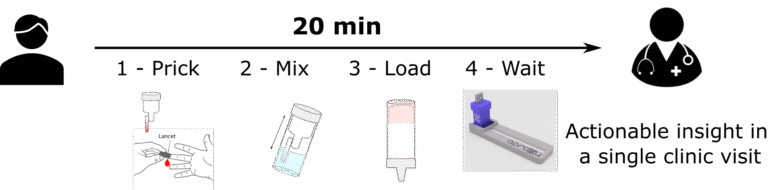
References
- Y.S. Ang, L. Y. L. Yung, “DNA-Programmed Reaction to Evaluate Specific IgE for Allergy Point-of-Care Testing,” Small 21, 2500575 (2025) DOI: 10.1002/smll.202500575
- R. Lu, Y. S. Ang, K. Cheung, K. Y. Quek, W. Sin, E. Lee, S. L. Lim, L. Y. L. Yung, M. E. Birnbaum, J. Han, L. F. Cheow, Z. K. Kwek, “iSECRETE: Integrating Microfluidics and DNA Proximity Amplification for Synchronous Single-Cell Activation and IFN-𝜸 Secretion Profiling,” Adv. Sci. 11, 2309920 (2024) DOI: 10.1002/advs.202309920
- Y. S. Ang and L. Y. L. Yung, “Protein-to-DNA Converter with High Signal Gain,” ACS Nano 18, 10454-10463 (2024) DOI: https://doi.org/10.1021/acsnano.3c11435
- Y. S. Ang, X. Qiu, H. M. Yam, N. Wu, L. Y. L. Yung, “Enzyme-free and isothermal discrimination of microRNA point mutations using a DNA split proximity circuit with turn-on fluorescence readout,” Biosens. Bioelectron. 214, 114727 (2022) DOI: https://doi.org/10.1016/j.bios.2022.114727
- Y. S. Ang and L. Y. L. Yung, “Dynamically elongated associative toehold for tuning DNA circuit kinetics and thermodynamics,” Nucleic Acids Res. 49, 4258-4265 (2021) DOI: https://doi.org/10.1093/nar/gkab212
- Y. S. Ang, P. S. Lai, L. Y. L. Yung, “Design of Split Proximity Circuit as a Plug-and-Play Translator for Point Mutation Discrimination,” Anal. Chem. 92, 11164-11170 (2020) DOI: 10.1021/acs.analchem.0c01379
- Y. S. Ang, J. J. Li, P. J. Chua, C. T. Ng, B. H. Bay, L. Y. L. Yung, “Localized Visualization and Autonomous Detection of Cell Surface Receptor Clusters Using DNA Proximity Circuit,” Anal. Chem., 90, 6193-6198 (2018) DOI: 10.1021/acs.analchem.8b00722
- Y. S. Ang, R. Tong, L. Y. L. Yung, “Engineering a robust DNA split proximity circuit with minimized circuit leakage,” Nucleic Acids Res., 44, e121 (2016) DOI: 10.1093/nar/gkw447
- Y. S. Ang and L. Y. L. Yung, “Rational design of hybridization chain reaction monomers for robust signal amplification,” Chem. Commun., 52, 4219-4222 (2016) DOI: 10.1039/C5CC08907G